Introduction
The actin cytoskeleton is an essential intracellular filamentous structure that underpins cellular transport and cytoplasmic streaming in plant cells. Yet, the system-level properties of actin-based cellular trafficking remain tenuous, largely due to the inability to quantify key features of the actin cytoskeleton. Here, we developed an automated image-based, network-driven framework to accurately segment and quantify actin cytoskeletal structures and Golgi transport. We show that the actin cytoskeleton in both growing and elongated hypocotyl cells has structural properties facilitating efficient transport. Our findings suggest that the erratic movement of Golgi is a stable cellular phenomenon that might optimize distribution efficiency of cell material. Moreover, we demonstrate that Golgi transport in hypocotyl cells can be accurately predicted from the actin network topology alone. Thus, our framework provides quantitative evidence for system-wide coordination of cellular transport in plant cells, and can be readily applied to investigate cytoskeletal organization and transport in other organisms.
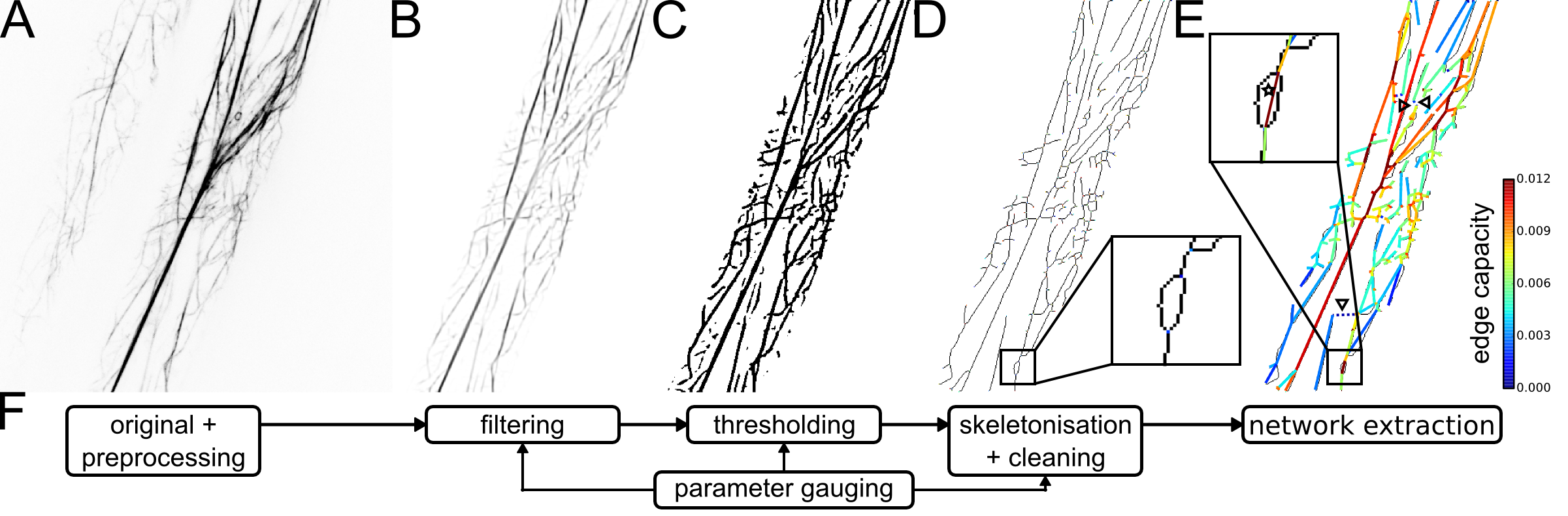
Extraction of networks from actin cytoskeletal image data.
(A) Gray-scale confocal image of two Arabidopsis hypocotyl cells after registration and background subtraction (“original + preprocessing”).
(B) Cytoskeleton image with improved signal-to-noise ratio after cropping of the largest cell and application of tubeness filter (“filtering”).
(C) Binary cytoskeleton image after application of adaptive median threshold (“thresholding”).
(D) Skeletonised cytoskeletal structures after removal of spurious fragments (“skeletonisation + cleaning”).
(E) Overlay of skeleton image and extracted cytoskeletal network with edges colour-coded by their capacity, reflecting the average filament thickness (“network extraction”).
(F) Overview of automated pipeline for extraction of actin cytoskeletal networks from image data.
Download
Download source-code (Fiji and Python) and example recording
Download complete, unprocessed imaging data (~500MB)
The software is available under GPL3.
Running the source-code requires the following packages: Python (2 or 3), SciPy, NumPy, Pandas, lxml, Shapely, NetworkX, and scikit-image
Instructions
Preprocessing of actin and Golgi image data:
- Download the Fiji source-code above (CytoSeg_ImagePreprocessing.py).
- Open and run the script using Fiji.
- Select the directory containing both actin and Golgi image data and adjust the file names in the script.
- Select the cellular region of interest when asked and confirm selection to proceed.
- The preprocessed images and tracking data are written to the selected directory.
Extraction of actin networks from image data:
- Download the Python source-code above (CytoSeg_NetworkExtraction.py and CytoSeg_HelpFunctions.py).
- Open the script and adjust parameters as needed.
- Run the script using Python (2 or 3).
- Exemplary plots and the computed network properties are written to the selected directory.
Citation
If this software is useful for your work, please include one of the following references in your publication or redistribution:
Breuer et al. (2017) System-wide organization of actin cytoskeleton determines organelle transport in plant hypocotyl cells. PNAS, 10.1073/pnas.1706711114.
Contact
David Breuer
Systems Biology and Mathematical Modeling Group
Max Planck Institute of Molecular Plant Physiology
Am Muehlenberg 1
D-14476 Potsdam-Golm
Germany
Last update: 2017-04-08
