img2net
Introduction
Automated analysis of imaged phenotypes enables fast and reproducible quantification of biologically relevant features. Despite recent developments, recordings of complex, networked structures, such as: leaf venation patterns, cytoskeletal structures, or traffic networks, remain challenging to analyze. Here we illustrate the applicability of img2net to automatedly analyze such structures by reconstructing the underlying network, computing relevant network properties, and statistically comparing networks of different types or under different conditions. The software can be readily used for analyzing image data of arbitrary 2D and 3D network-like structures.
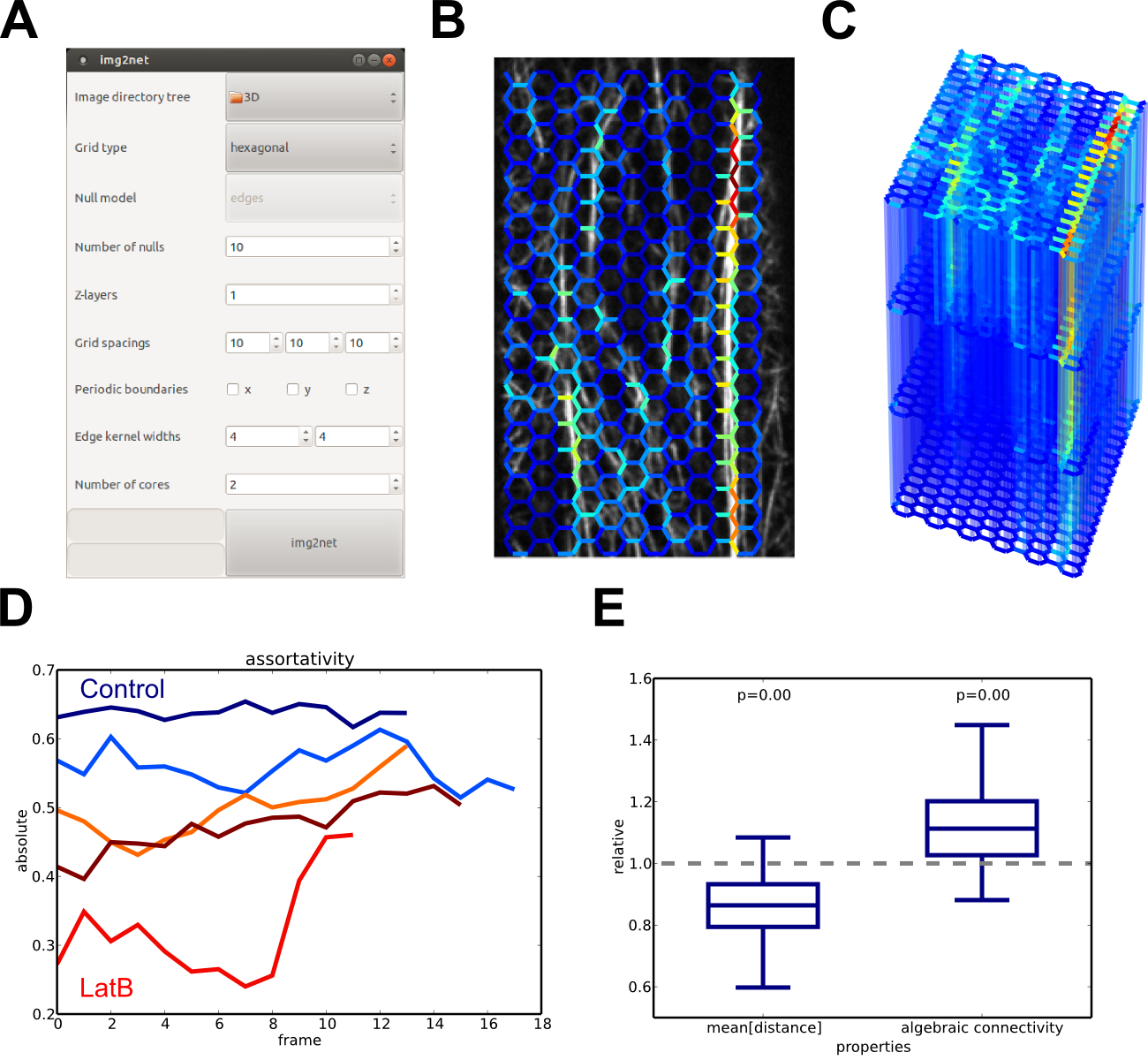
img2net: graphical user interface and output visualizations. (A) GUI to set parameters for the network reconstruction. (B) Actin network reconstructed with a hexagonal grid. (C) 3D reconstruction for image data with four z-slices. (D) Time series of the assortativity for multiple untreated (blue) and treated (red) plants. (E) Ratios of average path lengths and algebraic connectivities of observed and null model networks.
Download
Download binaries, single (Linux)
Download binaries, folder (Linux)
Download executable (Windows)
Download source-code (Python)
Download test data sets
The software is available under GPL3.
Running the source-code requires the following packages: Python 2.7.3, PyGTK, SciPy, NumPy, ParallelPython, NetworkX.
Under Windows, these packages come conveniently along with the bundles PythonXY and PyGTK.
The binaries/executables have been tested under 32-bit versions of Linux (Ubuntu 12.04 and Mint 13) and Windows (7 and 8).
Citation
If this software is useful for your work, please include one of the following references in your publication or redistribution:
Breuer et al. (2014) Quantitative analyses of the plant cytoskeleton reveal underlying organizational principles. J. R. Soc. Interface, 11(97): 20140362.
Breuer and Nikoloski (2014) img2net: Automated network-based analysis of imaged phenotypes. Bioinformatics, 30(22): 3291-3292.
Contact
David Breuer
Systems Biology and Mathematical Modeling Group
Max Planck Institute of Molecular Plant Physiology
Am Muehlenberg 1
D-14476 Potsdam-Golm
Germany
Last update: 2015-04-01
