
Introduction
Thread-like structures are pervasive across scales, from polymeric proteins to root systems to galaxy filaments, and their characteristics can be readily investigated in the network formalism. Yet, network links usually represent only parts of filaments, which, when neglected, may lead to erroneous conclusions from network-based analyses. The existing alternatives to detect filaments in network representations require tuning of parameters over a large range of values and treat all filaments equally, thus, precluding automated analysis of diverse filamentous systems. Here, we propose a fully automated and robust optimisation-based approach to detect filaments of consistent intensities and angles in a given network. We test and demonstrate the accuracy of our solution with contrived, biological, and cosmic filamentous structures. In particular, we show that the proposed approach provides powerful automated means to study properties of individual actin filaments in their network context.
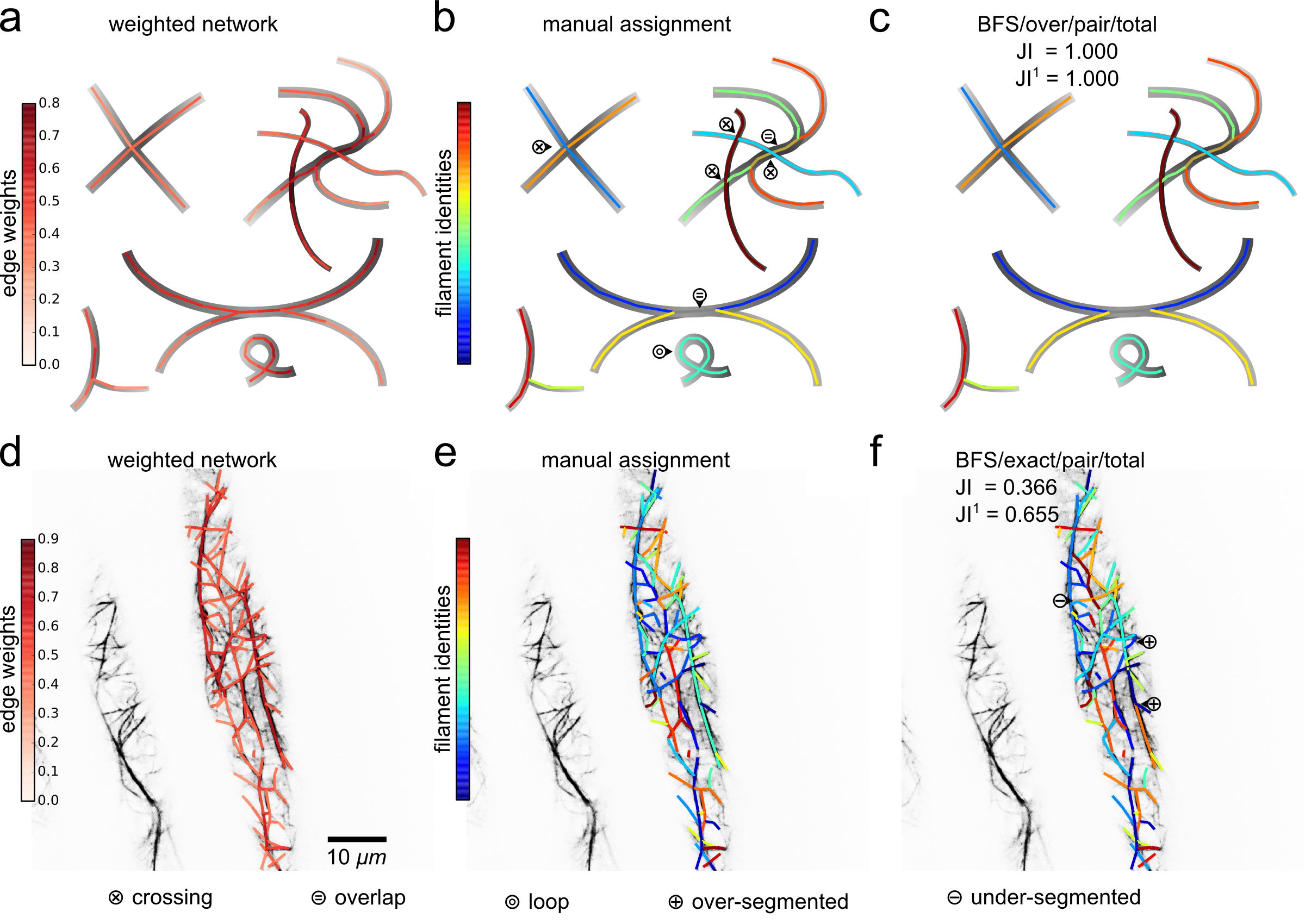
DeFiNe: Input networks, manual filament assignments, and automated filament decomposition from solving the filament cover optimization problem.
(A-C) Contrived filamentous network extracted from underlying drawing.
(D-E) Actin cytoskeletal network extracted from underlying confocal microscopy recording.
Download
Download binaries (Linux)
Download executable (Windows)
Download source-code (Python)
Download test data set
The software is available under GPL3.
Running the source-code requires the following packages: Python 2.7.3, PyGTK, SciPy, NumPy, NetworkX, GLPK, and CvxOpt.
Under Windows, these packages come conveniently along with the bundles PythonXY, PyGTK, and WinGLPK.
The binaries/executables have been tested under 32-bit versions of Linux (Ubuntu 12.04 and Mint 13) and Windows (7).
Citation
If this software is useful for your work, please include one of the following references in your publication or redistribution:
Breuer and Nikoloski (2015) DeFiNe: an optimisation-based method for robust disentangling of filamentous networks. Sci. Rep., 5:18267.
Contact
David Breuer
Systems Biology and Mathematical Modeling Group
Max Planck Institute of Molecular Plant Physiology
Am Muehlenberg 1
D-14476 Potsdam-Golm
Germany
Last update: 2014-09-07

