
Summary
Phytotyping4D is a non-invasive and accurate imaging system that combines a 3D light-field camera (Raytrix GmbH) with an automated pipeline, which provides validated measurements of growth, movement and other morphological features at the rosette and single-leaf level.
In a case study in which we investigated the link between starch and growth, we demonstrated that Phytotyping4D is a key step towards bridging the gap between phenotypic observations and the rich genetic and metabolic knowledge.
Our software was developed and optimized for greyscale 3D light-field images of Arabidopsis thaliana, and the parameters for rosette and leaf segmentation, leaf tracking and automated leaf number classification may be adapted to other plant species.
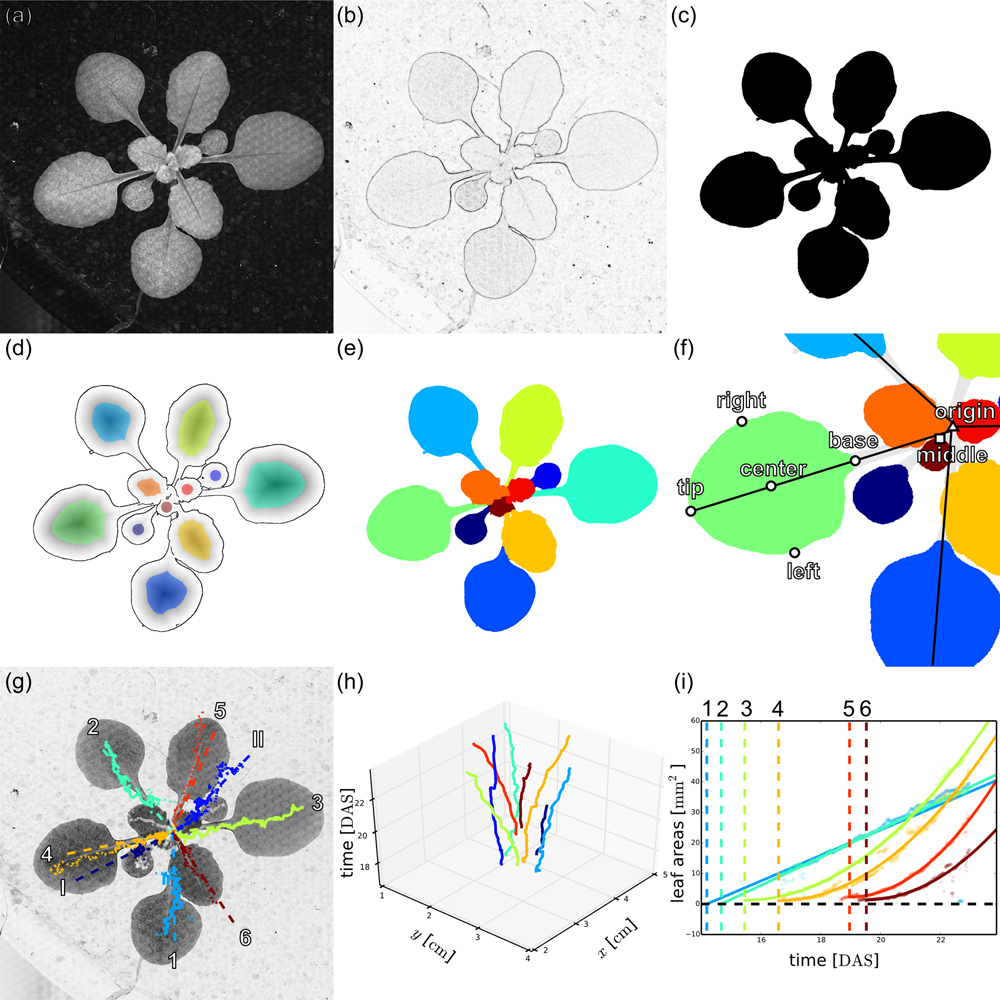
Phytotyping4D: Image processing pipeline for rosette and leaf segmentation and extraction of landmark points to analyse spatio-temporal growth.
(a) Original 2D focus image of an Arabidopsis Col-0 plant 24 days after sowing (DAS).
(b) Sobel edge filtered focus image.
(c) Watershed-based segmented rosette image.
(d) Overlay of the edge image from a Canny edge filter (lines), the Euclidean distance transform of the Canny edge image (grey-scale), and the seed points obtained from the local maxima of the distance transform (colours).
(e) Segmented leaves after watershed-based segmentation of the distance-transformed edge image.
(f) Scheme of rosette and leaf landmarks used to derive leaf properties.
(g) Automated leaf assignment on inverted focus image derived from the angular positions of the leaf tracks, assuming phyllotactic leaf angles follow the golden ratio (I and II: cotyledons, 1-6: first six leaves, numbered by their order of appearance).
(h) Three-dimensional plot of the tracked leaf centres over time, coloured according to leaf assignment.
(i) Timing of leaf appearance based on quadratic extrapolation of leaf area.
Download
Download source code (Python)
Download Col-0 test data set (created with RxLive software, version 2.8, Raytrix GmbH)
The software is available under GPL3.
Running the source code requires
Python and its packages
SciPy,
NumPy,
Matplotlib,
PyPng,
TrackPy, and
scikit-image.
The source code has been tested using Linux (Ubuntu 12.04).
Citation
If this software or part of it is useful for your work, please include one of the following references in your publication or redistribution:
Apelt et al. (2015) Phytotyping4D: A light-field imaging system for non-invasive and accurate monitoring of spatio-temporal plant growth. The Plant Journal, 82(4):693-706.
Apelt et al. (2017) Circadian, carbon, and light control of expansion growth and leaf movement. Plant Physiology, 174(2):00503.2017.
Contact
Federico Apelt and David Breuer
Max Planck Institute of Molecular Plant Physiology
Am Muehlenberg 1
D-14476 Potsdam-Golm
Germany
Last update: 2015-03-18

